Standardize metadata on-the-fly¶
This use cases runs on a LaminDB instance with populated CellType and Pathway registries. Make sure you run the GO Ontology notebook before executing this use case.
Here, we demonstrate how to standardize the metadata on-the-fly during cell type annotation and pathway enrichment analysis using these two registries.
For more information, see:
!lamin load use-cases-registries
💡 connected lamindb: testuser1/use-cases-registries
import lamindb as ln
import bionty as bt
from lamin_usecases import datasets as ds
import scanpy as sc
import matplotlib.pyplot as plt
import celltypist
import gseapy as gp
💡 connected lamindb: testuser1/use-cases-registries
sc.settings.set_figure_params(dpi=50, facecolor="white")
ln.settings.transform.stem_uid = "hsPU1OENv0LS"
ln.settings.transform.version = "0"
ln.track()
💡 notebook imports: bionty==0.43.1 celltypist==1.6.2 gseapy==1.1.3 lamin_usecases==0.0.1 lamindb==0.72.1 matplotlib==3.9.0 scanpy==1.10.1
💡 saved: Transform(uid='hsPU1OENv0LS6K79', version='0', name='Standardize metadata on-the-fly', key='analysis-registries', type='notebook', created_by_id=1, updated_at='2024-05-29 09:58:05 UTC')
💡 saved: Run(uid='S0jg8aslai7KQ6KRID8z', transform_id=1, created_by_id=1)
Run(uid='S0jg8aslai7KQ6KRID8z', started_at='2024-05-29 09:58:05 UTC', is_consecutive=True, transform_id=1, created_by_id=1)
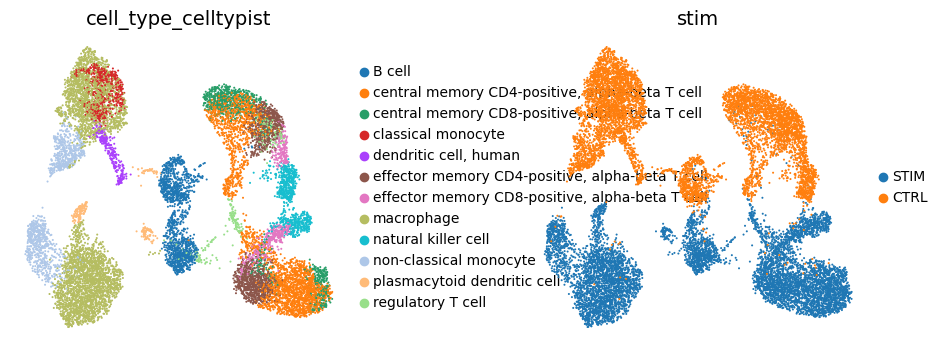
An interferon-beta treated dataset¶
A small peripheral blood mononuclear cell dataset that is split into control and stimulated groups. The stimulated group was treated with interferon beta.
Let’s load the dataset and perform some preprocessing:
adata = ds.anndata_seurat_ifnb(preprocess=False, populate_registries=True)
adata
AnnData object with n_obs × n_vars = 13999 × 9939
obs: 'stim'
var: 'symbol'
sc.pp.normalize_total(adata, target_sum=1e4)
sc.pp.log1p(adata)
sc.pp.highly_variable_genes(adata, n_top_genes=2000)
sc.pp.pca(adata, n_comps=20)
sc.pp.neighbors(adata, n_pcs=10)
sc.tl.umap(adata)
/opt/hostedtoolcache/Python/3.10.14/x64/lib/python3.10/site-packages/tqdm/auto.py:21: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html
from .autonotebook import tqdm as notebook_tqdm
Analysis: cell type annotation using CellTypist¶
model = celltypist.models.Model.load(model="Immune_All_Low.pkl")
Show code cell output
🔎 No available models. Downloading...
📜 Retrieving model list from server https://celltypist.cog.sanger.ac.uk/models/models.json
📚 Total models in list: 48
📂 Storing models in /home/runner/.celltypist/data/models
💾 Downloading model [1/48]: Immune_All_Low.pkl
💾 Downloading model [2/48]: Immune_All_High.pkl
💾 Downloading model [3/48]: Adult_CynomolgusMacaque_Hippocampus.pkl
💾 Downloading model [4/48]: Adult_Human_PancreaticIslet.pkl
💾 Downloading model [5/48]: Adult_Human_Skin.pkl
💾 Downloading model [6/48]: Adult_Mouse_Gut.pkl
💾 Downloading model [7/48]: Adult_Mouse_OlfactoryBulb.pkl
💾 Downloading model [8/48]: Adult_Pig_Hippocampus.pkl
💾 Downloading model [9/48]: Adult_RhesusMacaque_Hippocampus.pkl
💾 Downloading model [10/48]: Autopsy_COVID19_Lung.pkl
💾 Downloading model [11/48]: COVID19_HumanChallenge_Blood.pkl
💾 Downloading model [12/48]: COVID19_Immune_Landscape.pkl
💾 Downloading model [13/48]: Cells_Adult_Breast.pkl
💾 Downloading model [14/48]: Cells_Fetal_Lung.pkl
💾 Downloading model [15/48]: Cells_Human_Tonsil.pkl
💾 Downloading model [16/48]: Cells_Intestinal_Tract.pkl
💾 Downloading model [17/48]: Cells_Lung_Airway.pkl
💾 Downloading model [18/48]: Developing_Human_Brain.pkl
💾 Downloading model [19/48]: Developing_Human_Gonads.pkl
💾 Downloading model [20/48]: Developing_Human_Hippocampus.pkl
💾 Downloading model [21/48]: Developing_Human_Organs.pkl
💾 Downloading model [22/48]: Developing_Human_Thymus.pkl
💾 Downloading model [23/48]: Developing_Mouse_Brain.pkl
💾 Downloading model [24/48]: Developing_Mouse_Hippocampus.pkl
💾 Downloading model [25/48]: Fetal_Human_AdrenalGlands.pkl
💾 Downloading model [26/48]: Fetal_Human_Pancreas.pkl
💾 Downloading model [27/48]: Fetal_Human_Pituitary.pkl
💾 Downloading model [28/48]: Fetal_Human_Retina.pkl
💾 Downloading model [29/48]: Fetal_Human_Skin.pkl
💾 Downloading model [30/48]: Healthy_Adult_Heart.pkl
💾 Downloading model [31/48]: Healthy_COVID19_PBMC.pkl
💾 Downloading model [32/48]: Healthy_Human_Liver.pkl
💾 Downloading model [33/48]: Healthy_Mouse_Liver.pkl
💾 Downloading model [34/48]: Human_AdultAged_Hippocampus.pkl
💾 Downloading model [35/48]: Human_Colorectal_Cancer.pkl
💾 Downloading model [36/48]: Human_Developmental_Retina.pkl
💾 Downloading model [37/48]: Human_Embryonic_YolkSac.pkl
💾 Downloading model [38/48]: Human_IPF_Lung.pkl
💾 Downloading model [39/48]: Human_Longitudinal_Hippocampus.pkl
💾 Downloading model [40/48]: Human_Lung_Atlas.pkl
💾 Downloading model [41/48]: Human_PF_Lung.pkl
💾 Downloading model [42/48]: Lethal_COVID19_Lung.pkl
💾 Downloading model [43/48]: Mouse_Dentate_Gyrus.pkl
💾 Downloading model [44/48]: Mouse_Isocortex_Hippocampus.pkl
💾 Downloading model [45/48]: Mouse_Postnatal_DentateGyrus.pkl
💾 Downloading model [46/48]: Mouse_Whole_Brain.pkl
💾 Downloading model [47/48]: Nuclei_Lung_Airway.pkl
💾 Downloading model [48/48]: Pan_Fetal_Human.pkl
predictions = celltypist.annotate(
adata, model="Immune_All_Low.pkl", majority_voting=True
)
adata.obs["cell_type_celltypist"] = predictions.predicted_labels.majority_voting
🔬 Input data has 13999 cells and 9939 genes
🔗 Matching reference genes in the model
🧬 3696 features used for prediction
⚖️ Scaling input data
🖋️ Predicting labels
✅ Prediction done!
👀 Detected a neighborhood graph in the input object, will run over-clustering on the basis of it
⛓️ Over-clustering input data with resolution set to 10
🗳️ Majority voting the predictions
✅ Majority voting done!
adata.obs["cell_type_celltypist"] = bt.CellType.standardize(
adata.obs["cell_type_celltypist"]
)
Analysis: Pathway enrichment analysis using Enrichr¶
This analysis is based on the GSEApy scRNA-seq Example.
First, we compute differentially expressed genes using a Wilcoxon test between stimulated and control cells.
# compute differentially expressed genes
sc.tl.rank_genes_groups(
adata,
groupby="stim",
use_raw=False,
method="wilcoxon",
groups=["STIM"],
reference="CTRL",
)
rank_genes_groups_df = sc.get.rank_genes_groups_df(adata, "STIM")
rank_genes_groups_df.head()
| names | scores | logfoldchanges | pvals | pvals_adj | |
|---|---|---|---|---|---|
| 0 | ISG15 | 99.454422 | 7.132726 | 0.0 | 0.0 |
| 1 | ISG20 | 96.735252 | 5.074347 | 0.0 | 0.0 |
| 2 | IFI6 | 94.970428 | 5.828650 | 0.0 | 0.0 |
| 3 | IFIT3 | 92.481857 | 7.432406 | 0.0 | 0.0 |
| 4 | IFIT1 | 90.698891 | 8.053686 | 0.0 | 0.0 |
Next, we filter out up/down-regulated differentially expressed gene sets:
degs_up = rank_genes_groups_df[
(rank_genes_groups_df["logfoldchanges"] > 0)
& (rank_genes_groups_df["pvals_adj"] < 0.05)
]
degs_dw = rank_genes_groups_df[
(rank_genes_groups_df["logfoldchanges"] < 0)
& (rank_genes_groups_df["pvals_adj"] < 0.05)
]
degs_up.shape, degs_dw.shape
((541, 5), (933, 5))
Run pathway enrichment analysis on DEGs and plot top 10 pathways:
enr_up = gp.enrichr(degs_up.names, gene_sets="GO_Biological_Process_2023").res2d
gp.dotplot(enr_up, figsize=(2, 3), title="Up", cmap=plt.cm.autumn_r);
Register analyzed dataset and annotate with metadata¶
gRegister new features and labels (check out more details here):
new_features = ln.Feature.from_df(adata.obs)
ln.save(new_features)
new_labels = [ln.ULabel(name=i) for i in adata.obs["stim"].unique()]
ln.save(new_labels)
features = ln.Feature.lookup()
Register dataset using a Artifact object:
artifact = ln.Artifact.from_anndata(
adata,
description="seurat_ifnb_activated_Bcells",
)
artifact.save()
Artifact(uid='t1HzrAfm0gAaaULmK5rq', description='seurat_ifnb_activated_Bcells', suffix='.h5ad', accessor='AnnData', size=214787157, hash='5irB1DzU1RCyxFVygdWgUj', hash_type='sha1-fl', visibility=1, key_is_virtual=True, created_by_id=1, storage_id=1, transform_id=1, run_id=1, updated_at='2024-05-29 10:00:55 UTC')
artifact.features._add_set_from_anndata(
var_field=bt.Gene.symbol,
organism="human", # optionally, globally set organism via bt.settings.organism = "human"
)
Link cell type labels¶
cell_type_records = bt.CellType.from_values(adata.obs["cell_type_celltypist"])
artifact.labels.add(cell_type_records, features.cell_type_celltypist)
Link stimulation labels:
stim_records = ln.ULabel.from_values(adata.obs["stim"])
artifact.labels.add(stim_records, features.stim)
Link pathway labels¶
Let’s enable tracking of the current notebook as the transform of this artifact:
We further create two feature sets for degs_up and degs_dw which we can later associate with the associated pathways:
degs_up_featureset = ln.FeatureSet.from_values(
degs_up.names,
bt.Gene.symbol,
name="Up-regulated DEGs STIM vs CTRL",
type="category",
organism=( # optionally, globally set organism via bt.settings.organism = "human"
"human"
),
)
degs_dw_featureset = ln.FeatureSet.from_values(
degs_dw.names,
bt.Gene.symbol,
name="Down-regulated DEGs STIM vs CTRL",
type="category",
organism=( # optionally, globally set organism via bt.settings.organism = "human"
"human"
),
)
# Link feature sets to artifact
artifact.features.add_feature_set(degs_up_featureset, slot="STIM-up-DEGs")
artifact.features.add_feature_set(degs_dw_featureset, slot="STIM-down-DEGs")
Link the top 10 pathways to the corresponding differentially expressed genes:
def parse_ontology_id_from_enrichr_results(key):
"""Parse out the ontology id.
"ATF6-mediated Unfolded Protein Response (GO:0036500)" -> ("GO:0036500", "ATF6-mediated Unfolded Protein Response")
"""
id = key.split(" ")[-1].replace("(", "").replace(")", "")
name = key.replace(f" ({id})", "")
return (id, name)
# get ontology ids for the top 10 pathways
enr_up_top10 = [
pw_id[0]
for pw_id in enr_up.head(10).Term.apply(parse_ontology_id_from_enrichr_results)
]
enr_dw_top10 = [
pw_id[0]
for pw_id in enr_dw.head(10).Term.apply(parse_ontology_id_from_enrichr_results)
]
# get pathway records
enr_up_top10_pathways = bt.Pathway.from_values(enr_up_top10, bt.Pathway.ontology_id)
enr_dw_top10_pathways = bt.Pathway.from_values(enr_dw_top10, bt.Pathway.ontology_id)
Associate the pathways to the differentially expressed genes:
degs_up_featureset.pathways.set(enr_up_top10_pathways)
degs_dw_featureset.pathways.set(enr_dw_top10_pathways)
degs_up_featureset.pathways.list("name")
['defense response to symbiont',
'response to type II interferon',
'defense response to virus',
'negative regulation of viral genome replication',
'cellular response to cytokine stimulus',
'positive regulation of cytokine production',
'response to interferon-beta',
'negative regulation of viral process',
'response to cytokine',
'regulation of viral genome replication']
Querying metadata¶
artifact.describe()
Artifact(uid='t1HzrAfm0gAaaULmK5rq', description='seurat_ifnb_activated_Bcells', suffix='.h5ad', accessor='AnnData', size=214787157, hash='5irB1DzU1RCyxFVygdWgUj', hash_type='sha1-fl', visibility=1, key_is_virtual=True, updated_at='2024-05-29 10:00:56 UTC')
Provenance
.created_by = 'testuser1'
.storage = '/home/runner/work/lamin-usecases/lamin-usecases/docs/use-cases-registries'
.transform = 'Standardize metadata on-the-fly'
.run = '2024-05-29 09:58:05 UTC'
Labels
.cell_types = 'effector memory CD8-positive, alpha-beta T cell', 'B cell', 'effector memory CD4-positive, alpha-beta T cell', 'dendritic cell, human', 'macrophage', 'natural killer cell', 'classical monocyte', 'non-classical monocyte', 'plasmacytoid dendritic cell', 'regulatory T cell'
.ulabels = 'STIM', 'CTRL'
Features
'stim' = 'STIM', 'CTRL'
'cell_type_celltypist' = 'effector memory CD8-positive, alpha-beta T cell', 'B cell', 'effector memory CD4-positive, alpha-beta T cell', 'dendritic cell, human', 'macrophage', 'natural killer cell', 'classical monocyte', 'non-classical monocyte', 'plasmacytoid dendritic cell', 'regulatory T cell'
Feature sets
'var' = 'DENND4B', 'GLYR1', 'PLPP6', 'LBH', 'SRSF3', 'PFDN2', 'CYTL1', 'POLE3', 'COPG2', 'PTCD2', 'SEMA4D', 'CYREN', 'DIP2A', 'NUP155', 'ARF3', 'CDC14A', 'RCOR1', 'BRPF1', 'GALNT10', 'GNPTG'
'obs' = 'stim', 'cell_type_celltypist'
'STIM-up-DEGs' = 'RRAGC', 'ADTRP', 'ADAR', 'FAS', 'PLAAT2', 'SLAMF7', 'APOBEC3B', 'VAMP5', 'MAX', 'RAB29', 'RBCK1', 'ACOT9', 'PI4K2B', 'TMX1', 'CTSS', 'GBP3', 'UBXN2A', 'FCN1', 'BATF2', 'TNFAIP8'
'STIM-down-DEGs' = 'C5AR1', 'SRSF3', 'AURKAIP1', 'HMGN1', 'VPS28', 'PLK3', 'ITGB1', 'HDAC1', 'VKORC1', 'STRAP', 'ATP5PF', 'ENG', 'OLR1', 'SEC62', 'PSAP', 'RTN4', 'EREG', 'IFI27L2'
Querying cell types¶
Querying for cell types contains “B cell” in the name:
bt.CellType.filter(name__contains="B cell").df().head()
| uid | name | ontology_id | abbr | synonyms | description | public_source_id | run_id | created_by_id | updated_at | |
|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||
| 1 | ryEtgi1y | B cell | CL:0000236 | None | B lymphocyte|B-cell|B cells|B-lymphocyte|Cycli... | A Lymphocyte Of B Lineage That Is Capable Of B... | 29 | None | 1 | 2024-05-29 09:57:56.491573+00:00 |
| 2 | 2EhFTUoZ | follicular B cell | CL:0000843 | None | Follicular B cells|follicular B-lymphocyte|Fo ... | A Resting Mature B Cell That Has The Phenotype... | 29 | None | 1 | 2024-05-29 09:57:56.272333+00:00 |
| 3 | 4IowPafD | germinal center B cell | CL:0000844 | None | GC B lymphocyte|germinal center B lymphocyte|P... | A Rapidly Cycling Mature B Cell That Has Disti... | 29 | None | 1 | 2024-05-29 09:57:56.311170+00:00 |
| 4 | 2cUPBtY8 | memory B cell | CL:0000787 | None | memory B lymphocyte|Age-associated B cells|mem... | A Memory B Cell Is A Mature B Cell That Is Lon... | 29 | None | 1 | 2024-05-29 09:57:56.351173+00:00 |
| 5 | 3jdCg7zi | naive B cell | CL:0000788 | None | Naive B cells|naive B-cell|naive B lymphocyte|... | A Naive B Cell Is A Mature B Cell That Has The... | 29 | None | 1 | 2024-05-29 09:57:56.370589+00:00 |
Querying for all artifacts annotated with a cell type:
celltypes = bt.CellType.lookup()
celltypes.plasmacytoid_dendritic_cell
CellType(uid='3JO0EdVd', name='plasmacytoid dendritic cell', ontology_id='CL:0000784', synonyms='pDC|type 2 DC|plasmacytoid T cell|T-associated plasma cell|lymphoid dendritic cell|IPC|plasmacytoid monocyte|DC2|interferon-producing cell', description='A Dendritic Cell Type Of Distinct Morphology, Localization, And Surface Marker Expression (Cd123-Positive) From Other Dendritic Cell Types And Associated With Early Stage Immune Responses, Particularly The Release Of Physiologically Abundant Amounts Of Type I Interferons In Response To Infection.', created_by_id=1, public_source_id=29, updated_at='2024-05-29 09:57:49 UTC')
ln.Artifact.filter(cell_types=celltypes.plasmacytoid_dendritic_cell).df()
| uid | version | description | key | suffix | accessor | size | hash | hash_type | n_objects | n_observations | visibility | key_is_virtual | storage_id | transform_id | run_id | created_by_id | updated_at | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||||||||||
| 1 | t1HzrAfm0gAaaULmK5rq | None | seurat_ifnb_activated_Bcells | None | .h5ad | AnnData | 214787157 | 5irB1DzU1RCyxFVygdWgUj | sha1-fl | None | None | 1 | True | 1 | 1 | 1 | 1 | 2024-05-29 10:00:56.908002+00:00 |
Querying pathways¶
Querying for pathways contains “interferon-beta” in the name:
bt.Pathway.filter(name__contains="interferon-beta").df()
| uid | name | ontology_id | abbr | synonyms | description | public_source_id | run_id | created_by_id | updated_at | |
|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||
| 684 | 1l4z0v8W | cellular response to interferon-beta | GO:0035458 | None | cellular response to fibroblast interferon|cel... | Any Process That Results In A Change In State ... | 64 | None | 1 | 2024-05-29 09:56:33.339286+00:00 |
| 2130 | 1NzHDJDi | negative regulation of interferon-beta production | GO:0032688 | None | down regulation of interferon-beta production|... | Any Process That Stops, Prevents, Or Reduces T... | 64 | None | 1 | 2024-05-29 09:56:33.494615+00:00 |
| 3127 | 3x0xmK1y | positive regulation of interferon-beta production | GO:0032728 | None | positive regulation of IFN-beta production|up-... | Any Process That Activates Or Increases The Fr... | 64 | None | 1 | 2024-05-29 09:56:33.605094+00:00 |
| 4334 | 54R2a0el | regulation of interferon-beta production | GO:0032648 | None | regulation of IFN-beta production | Any Process That Modulates The Frequency, Rate... | 64 | None | 1 | 2024-05-29 09:56:33.738948+00:00 |
| 4953 | 3VZq4dMe | response to interferon-beta | GO:0035456 | None | response to fiblaferon|response to fibroblast ... | Any Process That Results In A Change In State ... | 64 | None | 1 | 2024-05-29 09:56:33.807058+00:00 |
Query pathways from a gene:
bt.Pathway.filter(genes__symbol="KIR2DL1").df()
| uid | name | ontology_id | abbr | synonyms | description | public_source_id | run_id | created_by_id | updated_at | |
|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||
| 1346 | 7S7qlEkG | immune response-inhibiting cell surface recept... | GO:0002767 | None | immune response-inhibiting cell surface recept... | The Series Of Molecular Signals Initiated By A... | 64 | None | 1 | 2024-05-29 09:56:33.409206+00:00 |
Query artifacts from a pathway:
ln.Artifact.filter(feature_sets__pathways__name__icontains="interferon-beta").first()
Artifact(uid='t1HzrAfm0gAaaULmK5rq', description='seurat_ifnb_activated_Bcells', suffix='.h5ad', accessor='AnnData', size=214787157, hash='5irB1DzU1RCyxFVygdWgUj', hash_type='sha1-fl', visibility=1, key_is_virtual=True, created_by_id=1, storage_id=1, transform_id=1, run_id=1, updated_at='2024-05-29 10:00:56 UTC')
Query featuresets from a pathway to learn from which geneset this pathway was computed:
pathway = bt.Pathway.filter(ontology_id="GO:0035456").one()
pathway
Pathway(uid='3VZq4dMe', name='response to interferon-beta', ontology_id='GO:0035456', synonyms='response to fiblaferon|response to fibroblast interferon|response to interferon beta', description='Any Process That Results In A Change In State Or Activity Of A Cell Or An Organism (In Terms Of Movement, Secretion, Enzyme Production, Gene Expression, Etc.) As A Result Of An Interferon-Beta Stimulus. Interferon-Beta Is A Type I Interferon.', created_by_id=1, public_source_id=64, updated_at='2024-05-29 09:56:33 UTC')
degs = ln.FeatureSet.filter(pathways__ontology_id=pathway.ontology_id).one()
Now we can get the list of genes that are differentially expressed and belong to this pathway:
contributing_genes = pathway.genes.all() & degs.genes.all()
contributing_genes.list("symbol")
['CALM1',
'IFITM1',
'PNPT1',
'OAS1',
'MNDA',
'AIM2',
'IFI16',
'IFITM3',
'STAT1',
'BST2',
'PLSCR1',
'SHFL',
'IFITM2',
'XAF1',
'IRF1']
# clean up test instance
!lamin delete --force use-cases-registries
!rm -r ./use-cases-registries
Show code cell output
Traceback (most recent call last):
File "/opt/hostedtoolcache/Python/3.10.14/x64/bin/lamin", line 8, in <module>
sys.exit(main())
File "/opt/hostedtoolcache/Python/3.10.14/x64/lib/python3.10/site-packages/rich_click/rich_command.py", line 367, in __call__
return super().__call__(*args, **kwargs)
File "/opt/hostedtoolcache/Python/3.10.14/x64/lib/python3.10/site-packages/click/core.py", line 1157, in __call__
return self.main(*args, **kwargs)
File "/opt/hostedtoolcache/Python/3.10.14/x64/lib/python3.10/site-packages/rich_click/rich_command.py", line 152, in main
rv = self.invoke(ctx)
File "/opt/hostedtoolcache/Python/3.10.14/x64/lib/python3.10/site-packages/click/core.py", line 1688, in invoke
return _process_result(sub_ctx.command.invoke(sub_ctx))
File "/opt/hostedtoolcache/Python/3.10.14/x64/lib/python3.10/site-packages/click/core.py", line 1434, in invoke
return ctx.invoke(self.callback, **ctx.params)
File "/opt/hostedtoolcache/Python/3.10.14/x64/lib/python3.10/site-packages/click/core.py", line 783, in invoke
return __callback(*args, **kwargs)
File "/opt/hostedtoolcache/Python/3.10.14/x64/lib/python3.10/site-packages/lamin_cli/__main__.py", line 103, in delete
return delete(instance, force=force)
File "/opt/hostedtoolcache/Python/3.10.14/x64/lib/python3.10/site-packages/lamindb_setup/_delete.py", line 98, in delete
n_objects = check_storage_is_empty(
File "/opt/hostedtoolcache/Python/3.10.14/x64/lib/python3.10/site-packages/lamindb_setup/core/upath.py", line 798, in check_storage_is_empty
raise InstanceNotEmpty(message)
lamindb_setup.core.upath.InstanceNotEmpty: Storage /home/runner/work/lamin-usecases/lamin-usecases/docs/use-cases-registries/.lamindb contains 1 objects ('_is_initialized' ignored) - delete them prior to deleting the instance
['/home/runner/work/lamin-usecases/lamin-usecases/docs/use-cases-registries/.lamindb/_is_initialized', '/home/runner/work/lamin-usecases/lamin-usecases/docs/use-cases-registries/.lamindb/t1HzrAfm0gAaaULmK5rq.h5ad']